Genomics of Climate Response in Plants
We are part of the Genomics of Thermotolerance in Tomato Reproduction consortium (NSF Plant Genomes Research Program IOS#1939255) to understand the genomic basis of heat-tolerance during pollen tube formation in tomato (Solanum). We are using a combination of population (and meta-population) genomic variation, RNA-Seq differential expression experimentation, and biochemical analyses to reveal this mechanism. We will also use wild population variation and environmental data to understand the ecological context of heat tolerance in the context of natural populations. Read past work in Molecular Ecology and PLoS Biology.
We are leading the African Acacia Eco-Evo-Genomics consortium (NSF Bridging Ecology and Evolution DEBS#2105917) to explore the combined effects of fire, drought, grass competition, and herbivory on the evolutionary history and present ecological responses in the African acacias (Vachellia and Senegalia; Fabaceae). We are using a combination of population genomics, phylogenetics, ecological common garden, and RNA-Seq differential expression analyses to explore questions about these fascinating savanna trees.


Vertebrate Muscle Genomes and Evolution of Extreme Muscle Behaviors
Muscles are a fundamental animal tissue critically involved in motility, circulation, digestion, reproduction. Despite their importance and a wealth of physiological comparative data, we still know little at the molecular evolutionary level about how muscle-related proteins have changed over the evolutionary history of vertebrate animals.
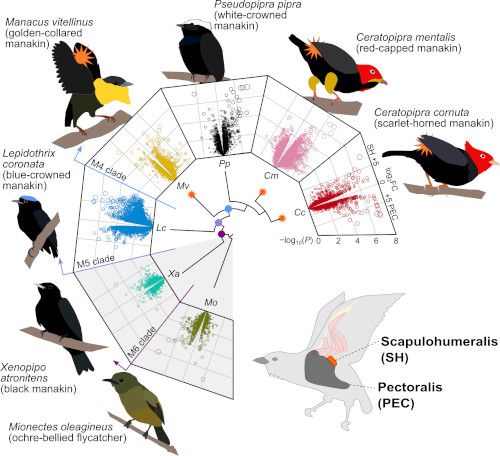
Our work focuses on understanding the diversification of muscle-related gene expression through evolutionary analysis of gene expression shifts over phylogenetic clades, alongside sequence-based analysis of selection, protein structural evolution, and gene family diversification. Recently, our work on understanding extreme muscle contractions related to mating displays in Manakins was features in Proceedings of the National Academy of Sciences. This work featured use of a new Phylogenetic Differential Gene Expresion Tool (PhyDGET)developed by our group.
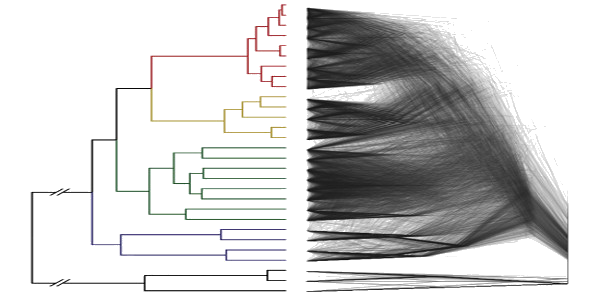
Population Genomics and Phylogenomics of Discordance
Comparative genomic data has revealed extensive gene flow, lineage sorting, and complex transcriptome expression patterns in a variety of systems. We are continuing to analytical approaches and computational methods to detect these patterns and how they overlap with speciation, complex trait adaptation, and patterns of sorting of ancestral variation.
Current work focuses on continued development of molecular phylogenetic methods for for detection of genomics discordance patterns, and expansion into understanding the relationship between molecular discordance and transcriptomic data. We are investigating these complex evolution histories largely in recently diverge groups, using a combination of population genomic, coalescent, and phylogenetic techniques. Read example publication in PLoS Biology and Science, Systematic Biology, and The American Journal of Botany.